Overview
cuCIM is a new RAPIDS library for accelerated n-dimensional image processing and image I/O. The project is now publicly available under a permissive license (Apache 2.0) and welcomes community contributions. In this post, we will cover the overall motivation behind the library and some initial benchmark results. In a complementary post on the Quansight blog, we provide guidelines on how existing CPU-based scikit-image code can be ported to the GPU. The library’s initial release was a collaboration between Quansight and the RAPIDS and Clara teams at NVIDIA.
Motivation
A primary objective of cuCIM is to provide open-source implementations of a wide range of CUDA-accelerated n-dimensional image processing operations that closely mirror the scikit-image API. Volumetric or general n-dimensional data is encountered in scientific fields such as bioimaging (microscopy), medical imaging (CT/PET/MRI), materials science, and remote sensing. This familiar, Pythonic API provides an accessible interface allowing researchers and data scientists to rapidly port existing CPU-based codes to the GPU.
Although a number of proprietary and open-source libraries provide GPU-accelerated 2D image processing operations (e.g. OpenCV, CUVI, VPI, NPP, DALI), these either lack 3D support or focus on a narrower range of operations than cuCIM. Popular n-dimensional image processing tools like scikit-image, SciPy’s ndimage module, and the Image Processing Toolkit (ITK and SimpleITK) have either no or minimal GPU support. The CLIJ library is an OpenCL-based 2D and 3D image processing library with some overlap in functionality with cuCIM. Although CLIJ is a Java-based project being developed among the ImageJ/Fiji community, the effort is underway to provide interfaces from Python and other languages (clEsperanto).
Aside from providing image processing functions, we also provide image/data readers designed to address the I/O bottlenecks commonly encountered in Deep Learning training scenarios. Both C++ and Python APIs for reading TIFF files with an API matching the OpenSlide library are provided, with support for additional image formats planned for future releases (e.g. DICOM, NIFTI, and the Zarr-based Next Generation File Format being developed by the Open Microscopy Environment (OME).
cuCIM Architecture
C++ Architecture and plugins
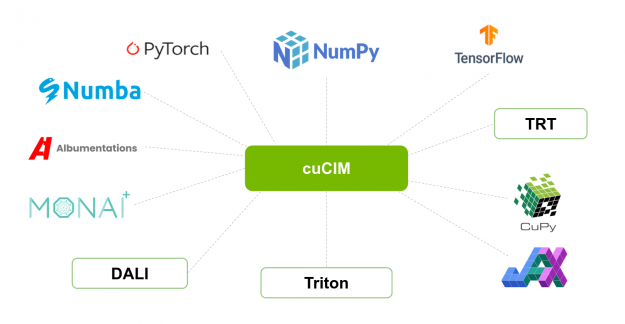
cuCIM consists of I/O, file system, and operation modules at its core layers. Image formats and operations on images supported by cuCIM can be extended through plugins. cuCIM’s plug-in architecture is based on NVIDIA Carbonite SDK which is a core engine of NVIDIA Omniverse applications.
The CuImage C++ class represents an image and its metadata. This CuImage class and functions in core modules such as TIFF loader and filesystem I/O using NVIDIA GPUDirect Storage (GDS — also known as cuFile) are also available from Python via the cucim.clara package
Pythonic image processing utilizing CuPy
The GPU-based implementation of the scikit-image API is provided in the cucim.skimage module. These functions have been implemented using the CuPy library. CuPy was chosen because it provides a GPU equivalent for most of NumPy and a substantial subset of SciPy (FFTs, sparse matrices, n-dimensional image processing primitives). By using technologies such as Thrust and CUB, efficient, templated sorting and reduction routines are available as well. For cases where custom CUDA kernels are needed, it also contains ElementwiseKernel and RawKernel classes that can be used to simplify the generation of the necessary kernels at run-time for the provided input data types.
Interoperability
Finally, CuPy supports both DLPack and the __cuda_array_interface__ protocols, making it possible to easily interoperate with other Python libraries with GPU support as Numba, PyCUDA, PyTorch, Tensorflow, JAX, and Dask. Other libraries in the RAPIDS ecosystem provide complementary functions covering areas such as machine learning (cuML), signal processing (cuSignal), or graph algorithms (cuGraph) that can be combined with cuCIM in applications. Additionally properties of ROIs (regions of interest) identified by an algorithm fit naturally in cuDF data-frames.
An example of using Dask to perform blockwise image processing was given in the following Dask deconvolution blog post, which illustrates distributed processing across multiple GPUs. Although the blog post was written prior to the release of cuCIM, the same approach of using Dask’s map_overlap function can be employed for many of the image processing operations in cuCIM. Blockwise processing (using map_blocks) can also be useful even with a single GPU in order to reduce peak GPU memory requirements.
Benchmarks
TIFF Image I/O
In the following figure, the relative performance for reading all non-overlapping (256, 256) patches from an RGB TIFF image of shape (81017, 92344, 3) on an SSD drive is shown. Performance is plotted in terms of acceleration factor relative to the OpenSlide library.
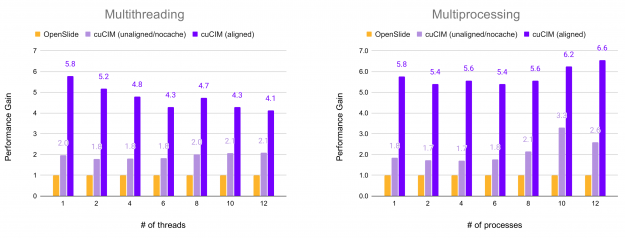
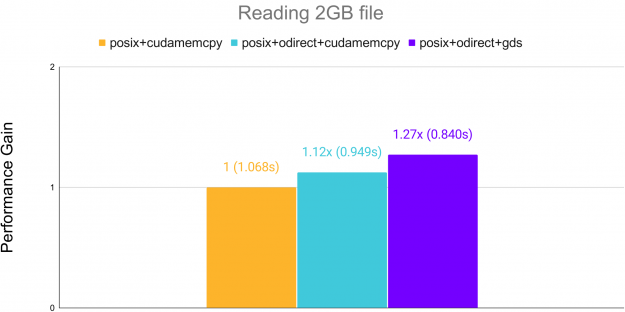
Additionally, reading raw binary data can see acceleration over standard cudaMemcpy when using GPUDirect Storage (GDS) on systems supporting it. GDS allows transfer directly from memory to the GPU, bypassing the CPU.
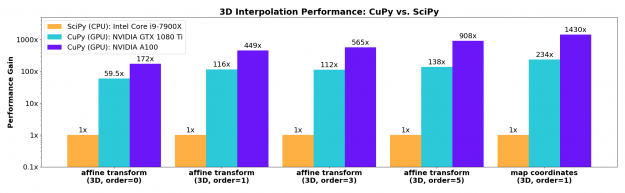
n-dimensional B-spline image interpolation
The necessary n-dimensional image interpolation routines needed to enable resizing, rotation, affine transforms, and warping were contributed upstream to CuPy’s cupyx.scipy.ndimage module. These implementations match the improved spline interpolation functions developed for SciPy 1.6 and support spline orders from 0-5 with a range of boundary conditions. These functions show excellent acceleration on the GPU, even for older generation gaming-class cards such as the NVIDIA GTX 1080-Ti.
Low-level operations
Image processing primitives such as filtering, morphology, color conversions, and labeling of connected components in binary images see substantial acceleration on the GPU. Performance numbers are in terms of acceleration factors relative to scikit-image 0.18.1 on the CPU.
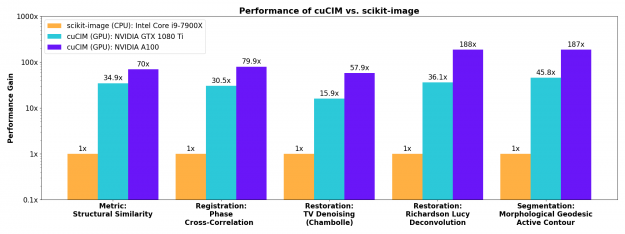
The results shown here are for relatively large images (2D images at 4K resolution). For smaller images such as (512, 512), acceleration factors will be smaller but still substantial. For tiny images (e.g. (32, 32)), processing on the CPU will be faster as there is some overhead involved in launching CUDA kernels on the GPU and there will not be enough work to keep all GPU cores busy.
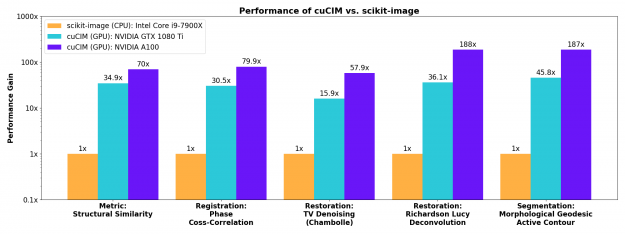
Higher-level operations
More involved image restoration, registration, and segmentation algorithms also see substantial acceleration on the GPU.
Contributing
cuCIM is an open-source project that welcomes community contributions!
Please visit Quansight’s blog to learn more about converting your CPU-based code to the GPU with cuCIM, and some areas on our roadmap where we would love to hear your feedback.