AWS for Industries
Easily run NVIDIA Parabricks Ready2Run workflows on Amazon Omics
Blog is guest authored by Harry Clifford from NVIDIA. To help customers easily build, deploy, and scale workloads, Amazon Omics now supports pre-built Ready2Run workflows from third-party software companies and open-source pipelines. Read more about the launch here.
As the cost of sequencing a human genome continues to decrease, the volume of sequencing data is exponentially increasing. Sequencing an individual’s whole genome generates roughly 100 gigabytes of raw data directly off a genome sequencing instrument. Many genome analysis pipelines are struggling to keep up with the expansive levels of raw data being generated at high speed and in high volumes, leading to a growing need for low-cost, accelerated analysis pipelines. Whether used for sequencing critical-care patients with rare diseases or in population-scale genetics research, whole genome sequencing is becoming a fundamental step in clinical workflows and drug discovery.
NVIDIA Parabricks is a suite of accelerated genomic analysis applications that contains optimized and AI-based industry-standard genomic tools such as GPU-accelerated GATK and DeepVariant. Parabricks can deliver accelerated analysis over CPU-based tools and reduce compute costs by utilizing NVIDIA GPUs. Thirteen Parabricks germline and somatic workflows are now available on Amazon Omics as Ready2Run workflows. Ready2Run workflows are a set of pre-built workflows from third-party software companies and open-source pipelines. With just a few clicks or a single API call, customers can run pre-built pipelines. Ready2Run workflows are priced-per-run to give customers predictable pricing.
NVIDIA has collaborated with various teams including the GATK team at the Broad Institute, to validate accuracy, producing results that are functionally equivalent to the CPU-native GATK versions.
Parabricks Workflows on Amazon Omics:
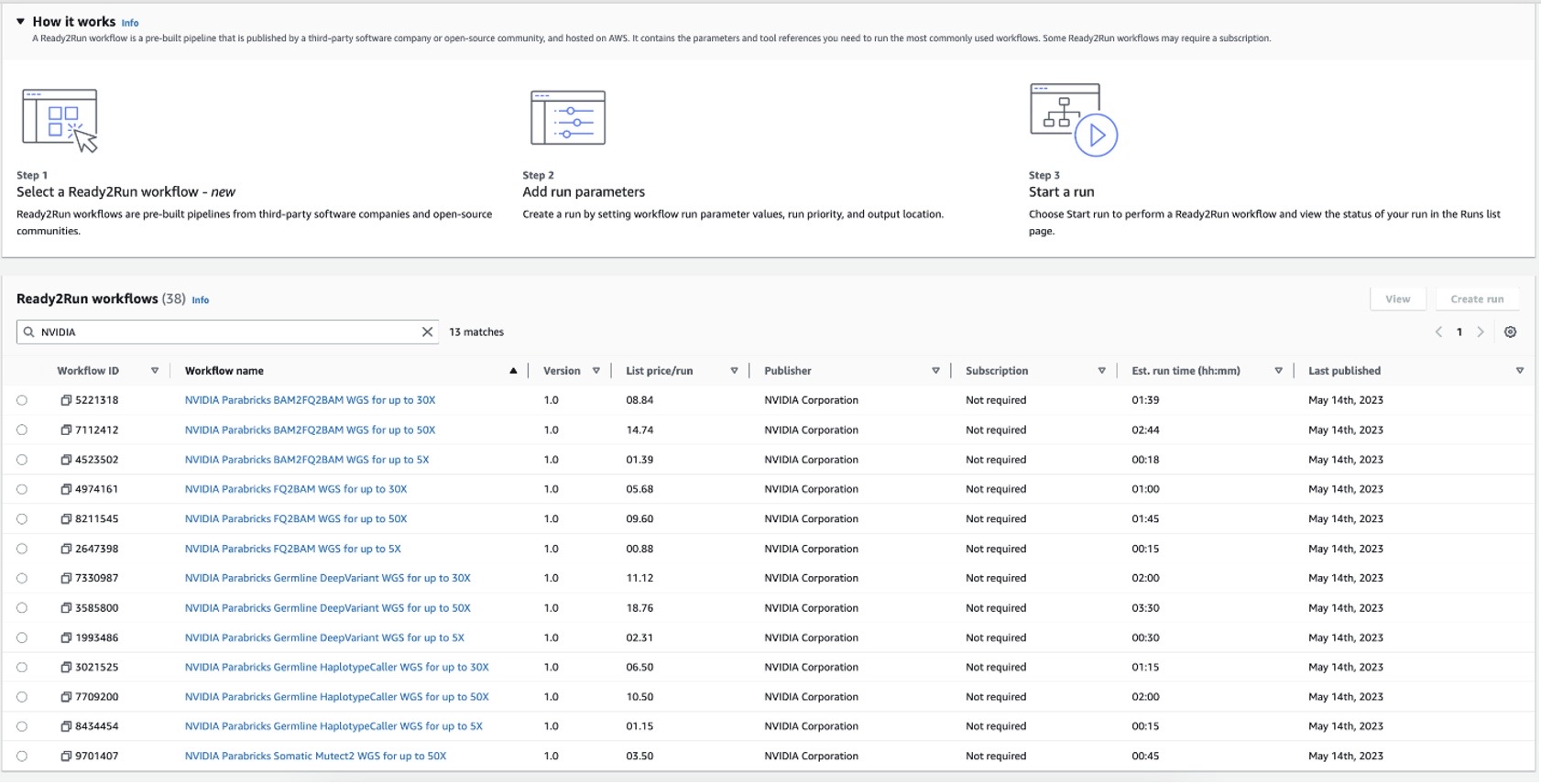 Figure 1: NVIDIA Ready2Run workflows on Amazon Omics displaying list price per run and estimated run time.
Figure 1: NVIDIA Ready2Run workflows on Amazon Omics displaying list price per run and estimated run time.
There is no license requirement to run the Parabricks workflows. This open-access policy aligns with NVIDIA’s goal to democratize accelerated genomics analysis and enable researchers around the world to replicate results achieved using Parabricks analysis workflows. For users who would like to have enterprise-level support, NVIDIA offers NVIDIA AI Enterprise.
With NVIDIA AI Enterprise, organizations receive full access to enterprise support that provides guaranteed response times, priority security notification, and access to Parabricks experts to trouble-shoot and optimize genomics workflows. NVIDIA AI Enterprise is designed to accelerate and streamline development and deployment.
Supported Pipelines:
The Parabricks Ready2Run workflows provide solutions for alignment, germline variant calling, somatic variant calling, and re-alignment to new reference genomes. Runtimes and costs for each workflow are transparent and predictable. Additionally, every workflow is pre-configured and tested, so no additional setup is needed to get started.
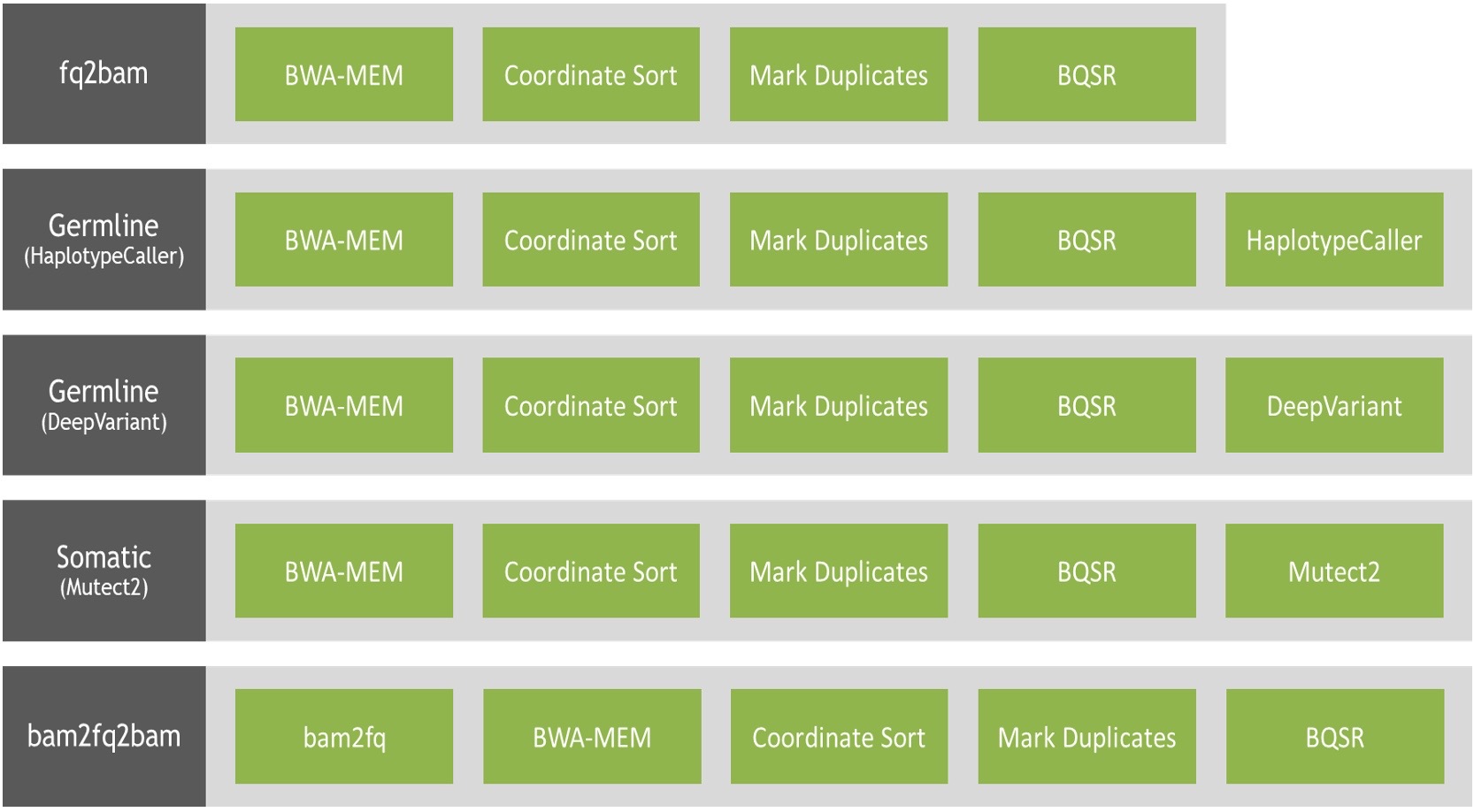 Figure 2: The 13 NVIDIA Parabricks Ready2Run workflows available on Amazon Omics span 5x, 30x, and 50x germline workflows for DeepVariant and HaplotypeCaller as well as a 50x somatic workflow.
Figure 2: The 13 NVIDIA Parabricks Ready2Run workflows available on Amazon Omics span 5x, 30x, and 50x germline workflows for DeepVariant and HaplotypeCaller as well as a 50x somatic workflow.
Alignment
The FQ2BAM workflow generates BAM/CRAM output given one or more pairs of FASTQ files, providing an accelerated version of BWA-MEM and pre-processing tools used in GATK4 best practices. This workflow provides functionally equivalent results to the CPU-native versions, but can align a 30X genome in one hour on Amazon Omics.
Germline
There are germline workflows for HaplotypeCaller and DeepVariant in Parabricks which generate VCFs as output. The germline (HaplotypeCaller) for WGS uses HaplotypeCaller and replicates GATK4 best practices, providing functionally equivalent results for a 30X whole genome on Amazon Omics.
The germline DeepVariant workflow for WGS utilizes an accelerated version of DeepVariant, a deep learning-based variant caller which provides increased accuracy of calls. DeepVariant is an AI-based model that is based on a CNN architecture and can be retrained on data for enhanced accuracy with each genomic platform or sequencing lab’s outputs. There are DeepVariant models available to choose from for multiple sequencing instruments, and this workflow can analyze a 30X whole genome on Amazon Omics.
Somatic
The Parabricks WGS 50x somatic workflow processes the tumor FASTQ files, and optionally normal FASTQ files and knownSites files, and generates tumor or tumor/normal analysis. The output is in VCF format. This workflow utilizes mutect2 and replicates GATK4 best practices for somatic analysis, and can align and variant call from a pair of deeper sequenced (50X) whole genomes on Amazon Omics.
Re-Alignment
The BAM2FQ2BAM workflow can be used to extract reads and realign to new reference genomes (such as the T2T completed human genome). This process of re-aligning has typically been very slow and computationally expensive, but with the Parabricks accelerated workflow, speedups of 10X are achieved.
This workflow un-aligns a BAM file, reversing it from BAM to FASTQ format, and realigns the FASTQ to produce a new BAM file on a different reference.
Conclusion
NVIDIA Parabricks Ready2Run workflows give customers the ability to easily run accelerated and AI-driven pipelines in the cloud.
To get started with NVIDIA Parabricks Ready2Run workflows, visit the Amazon Omics console.
To learn more about the price for each workflow, visit Amazon Omics Ready2Run pricing.
To learn more about enterprise support for NVIDIA Parabricks through NVIDIA AI Enteprise, contact NVIDIA.
Authors
Harry Clifford is the NVIDIA genomics product lead, leveraging NVIDIA’s expertise in AI, high performance computing (HPC), and data analytics stacks to address genomics workflows with accelerated high-accuracy solutions. His background is in bioinformatics and functional genomics, including a PhD from the University of Oxford, post-doctoral experience in the biopharma industry and at the University of Cambridge, and entrepreneurial experience in the biotech sector.